Clinical Utility
The traditional Rai and Binet clinical CLL staging systems are based on disease burden and have been useful for assigning patients to groups having similar survival times.1,2 These systems, however, are not effective in predicting survival in early-stage disease when most CLL cases are diagnosed. This has led to development of molecular markers that differentiate those patients who are prone to rapid progression from those who have indolent disease.
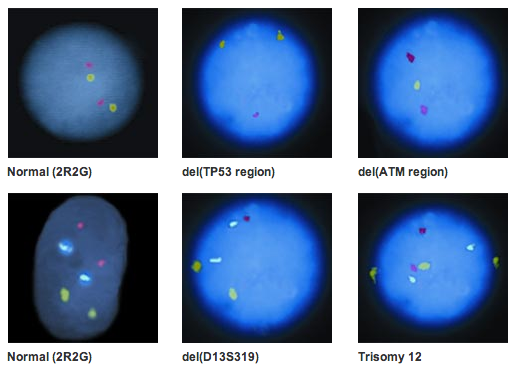
In a pivotal study conducted by Döhner et al, titled “Genomic Aberrations and Survival in Chronic Lymphocytic Leukemia,” genomic alterations as determined by FISH were found to be predictive for disease progression and overall survival.3 Multiple studies support the conclusion of Döhner et al. that loss of 17p and of 11q markers predicts reduced survival times as compared to other Döhner groups as determined by FISH aberrations.4,5,6
Such studies have led to the inclusion of FISH testing in the NCCN practice guidelines as a means to determine CLL prognosis. CLL prognostic categories according to the current NCCN guidelines divide patients into 3 categories – favorable, neutral and unfavorable.7
In a 2006 prospective study of 151 patients by Shanafelt et al utilizing Vysis FISH probes, a correlation was established between overall survival and FISH risk category for CLL at diagnosis.6 Patients were divided into 2 prognostic groups. They were assigned to the good/intermediate FISH prognosis group if there were no chromosomal aberrations or if only 13q-and/or +12 aberrations were present. If a chromosomal aberration of 17por 11q- was present, the patient was placed in the poor FISH prognosis group. Poor vs. good/intermediate FISH (P=0.004), age at diagnosis (P=0.0006); and Rai stage (P=0.0026) were each significantly associated with overall survival from diagnosis in univariate analysis. When all factors were included in multivariable Cox regression model, each of three factors still remained significant: Poor vs. good/intermediate FISH (P=0.00022), age at diagnosis (P=0.000024); and Rai stage (P=0.00012).6
The clinical utility of the Vysis CLL FISH Probe Kit has been established primarily from its high concordance with the assay employed in the publication by Shanafelt et al.6
Also, as noted in the Shanafelt study, all patients with the 17p- abnormality had between 24 to 94% of cells with this abnormality.6 Therefore, the effect of 17p- at very low levels could not be determined. In a publication on untreated 17p- CLL patients, Tam et al reported a 3-year overall survival of 92% for patients with < 25% 17p-deleted nuclei, vs. 54% for patients with ≥ 25% 17p-deleted nuclei (P=0.007).8
Method Concordance
A study was done comparing the method used in the Vysis CLL FISH Probe Kit and the method used by Shanafelt et al.6 This study established the clinical validity of the Vysis CLL FISH Probe Kit by demonstrating concordance to the FISH method used in the Shanafelt study. The clinical validity of the Vysis CLL FISH Probe Kit is documented via peer-reviewed literature.
The study analyzed 64 specimens whose Döhner classifications were based on previous results using the Shanafelt FISH method. Table 22 shows the distribution of specimens analyzed, by Döhner classification.